The covid19sf_geo provides information about San Francisco Covid19 cases distribution by geospatial location. Also, testing locations across San Francisco available on covid19sf_test_loc dataset. The following vignette provides examples for geospatial visualization of those datasets. Both datasets are sf objects and contain geometric information (i.e., ready to plot on a map).
Note: This is a non-CRAN vignette, and the following libraries required to build the plots on this document:
The covid19sf_geo dataset
The covid19sf_geo dataset provides a snapshot of the distribution of the Covid19 cases in San Francisco by different geographic locations splits of the city. The dataset contains the following fields:
-
area_type- the geograpichal split method:-
ZCTAfor view the data by ZIP code -
Analysis Neighborhoodfor view the data by neigborhoods -
Census Tractfor view the data by census tract, and -
Citywidefor total cases in the city
-
-
id- the area ID (e.g., ZIP code, neighborhood name, etc.) -
count- total number of positive cases in the area -
rate- cases rate per 10000 residents -
deaths- total number of deaths in the area -
acs_population- total number of residents in the area -
last_updated- most recent update time of the dataset
While the first three geographical split methods contain geometry components that enable us to plot them as a map, the last is just an aggregated summary of the city’s total cases.
library(covid19sf)
data(covid19sf_geo)
class(covid19sf_geo)
#> [1] "sf" "data.frame"
head(covid19sf_geo)
#> Simple feature collection with 6 features and 7 fields
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: -122.4543 ymin: 37.70792 xmax: -122.357 ymax: 37.80602
#> Geodetic CRS: WGS 84
#> area_type id count rate deaths
#> 1 Analysis Neighborhood Bayview Hunters Point 5314 1401.4822 37
#> 2 Analysis Neighborhood Bernal Heights 1729 687.0108 NA
#> 3 Analysis Neighborhood Castro/Upper Market 1211 538.1744 15
#> 4 Analysis Neighborhood Chinatown 561 388.5580 17
#> 5 Analysis Neighborhood Excelsior 3347 839.2888 27
#> 6 Analysis Neighborhood Financial District/South Beach 1309 607.7912 NA
#> acs_population last_updated geometry
#> 1 37917 2021-12-14 08:30:14 MULTIPOLYGON (((-122.3936 3...
#> 2 25167 2021-12-14 08:30:14 MULTIPOLYGON (((-122.4036 3...
#> 3 22502 2021-12-14 08:30:14 MULTIPOLYGON (((-122.4266 3...
#> 4 14438 2021-12-14 08:30:14 MULTIPOLYGON (((-122.4062 3...
#> 5 39879 2021-12-14 08:30:14 MULTIPOLYGON (((-122.425 37...
#> 6 21537 2021-12-14 08:30:14 MULTIPOLYGON (((-122.3875 3...Plotting cases with mapview
The most intuitive method for plotting sf objects is with the mapview package, which is a wrapper for the leaflet JavaScript package. The main advantage of the mapview package that it is both interactive and smoothly works with sf objects. The following example demonstrated the use case of the mapview function to plot the confirmed cases in San Francisco with the plot function to plot cases distribution by ZIP code:
You can use at and col.regions arguments to define color buckets and color range, respectively:
Plotting vaccine data with tmap
The tmap package provides functions and tools for creating thematic maps. The package supports sf objects and follow the ggplot2 framework. In the example below we will plot the COVID-19 vaccine data by geographic using the covid19sf_vaccine_geo dataset:
data(covid19sf_vaccine_geo)
head(covid19sf_vaccine_geo)
#> Simple feature collection with 6 features and 8 fields
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: -122.4773 ymin: 37.73155 xmax: -122.3843 ymax: 37.80602
#> Geodetic CRS: WGS 84
#> id area_type count_vaccinated_by_dph
#> 1 Bernal Heights Analysis Neighborhood 5106
#> 2 Financial District/South Beach Analysis Neighborhood 1841
#> 3 Glen Park Analysis Neighborhood 573
#> 4 Haight Ashbury Analysis Neighborhood 823
#> 5 Hayes Valley Analysis Neighborhood 2401
#> 6 Inner Sunset Analysis Neighborhood 1220
#> count_vaccinated count_series_completed acs_population
#> 1 21109 19781 25167
#> 2 22782 20215 21537
#> 3 7257 6804 8651
#> 4 14360 13279 19275
#> 5 16351 14930 19711
#> 6 23414 21829 29539
#> percent_pop_series_completed last_updated
#> 1 0.7859896 2021-12-15 04:45:07
#> 2 0.9386173 2021-12-15 04:45:09
#> 3 0.7864987 2021-12-15 04:45:09
#> 4 0.6889235 2021-12-15 04:45:09
#> 5 0.7574451 2021-12-15 04:45:09
#> 6 0.7389891 2021-12-15 04:45:09
#> geometry
#> 1 MULTIPOLYGON (((-122.4036 3...
#> 2 MULTIPOLYGON (((-122.3875 3...
#> 3 MULTIPOLYGON (((-122.4474 3...
#> 4 MULTIPOLYGON (((-122.432 37...
#> 5 MULTIPOLYGON (((-122.4208 3...
#> 6 MULTIPOLYGON (((-122.4529 3...Additional setting: Following changes in the default options of the sf package from version 1.0-1 by default option is to use s2 spherical geometry as default when coordinates are ellipsoidal. That cause some issues with the tmap package, therefore we will set this functionality as FALSE:
sf_use_s2(FALSE)
#> Spherical geometry (s2) switched offWe will use the percent_pop_series_completed variable to plot the percentage of population that finished their vaccination process. Let’s first filter the data and transform the percent_pop_series_completed from decimal to percentage:
df <- covid19sf_vaccine_geo %>% filter(area_type == "Analysis Neighborhood") %>%
dplyr::mutate(perc_complated = percent_pop_series_completed * 100)Now we can plot the new object:
tm_shape(df) +
tm_polygons("perc_complated",
title = "% Group")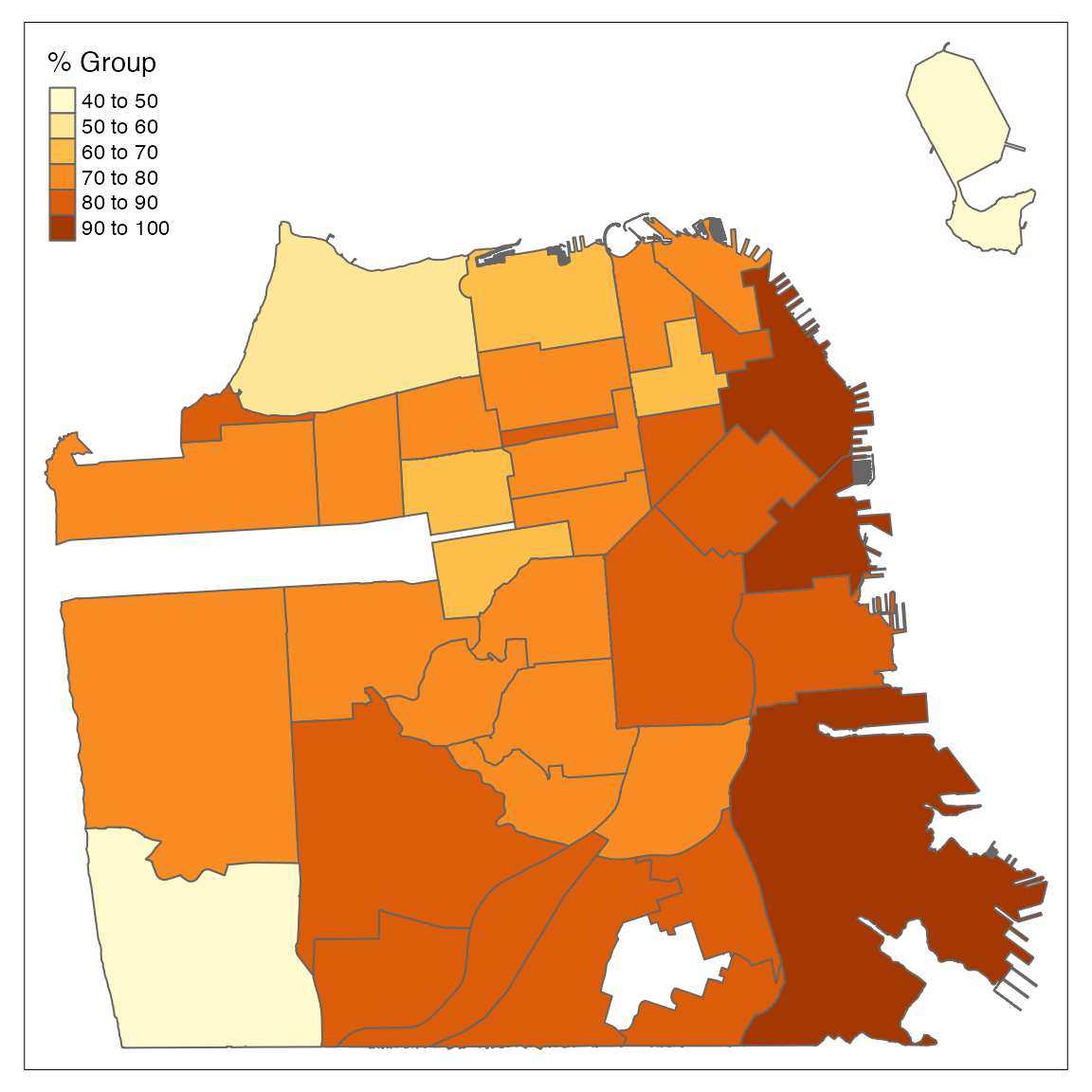
By default, the tm_polygons function bucket the numeric variable, in this case percentage of vaccinated population, into buckets. You can control the number of buckets using the n argument. Let’s now start to customize the plot and modify to color palette:
tm_shape(df) +
tm_polygons("perc_complated",
title = "% Group",
palette = "RdYlBu")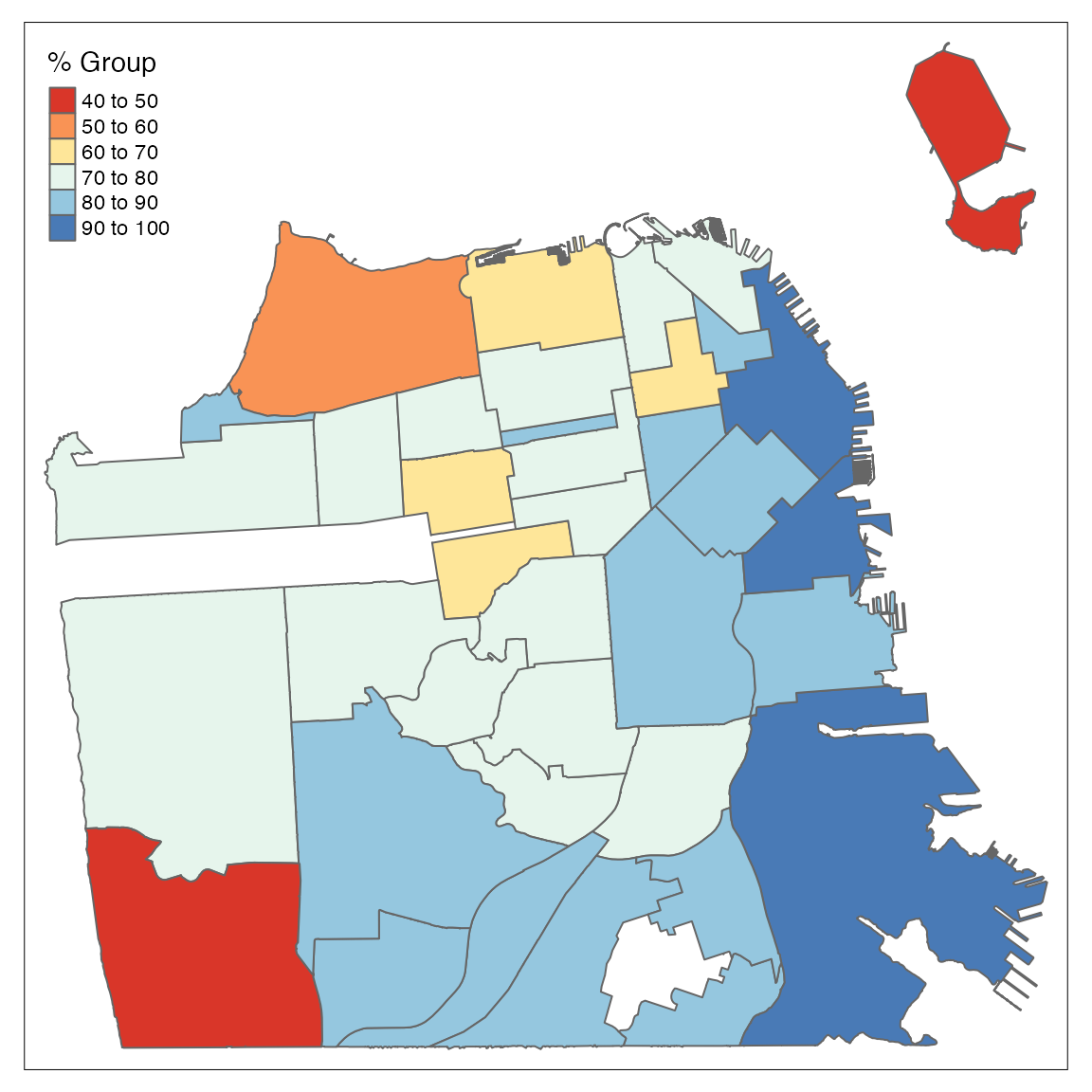
We can customize the plot background with the tm_style function by using the style argument:
tm_shape(df) +
tm_polygons("perc_complated",
title = "% Group",
palette = "RdYlBu") +
tm_style(style = "cobalt")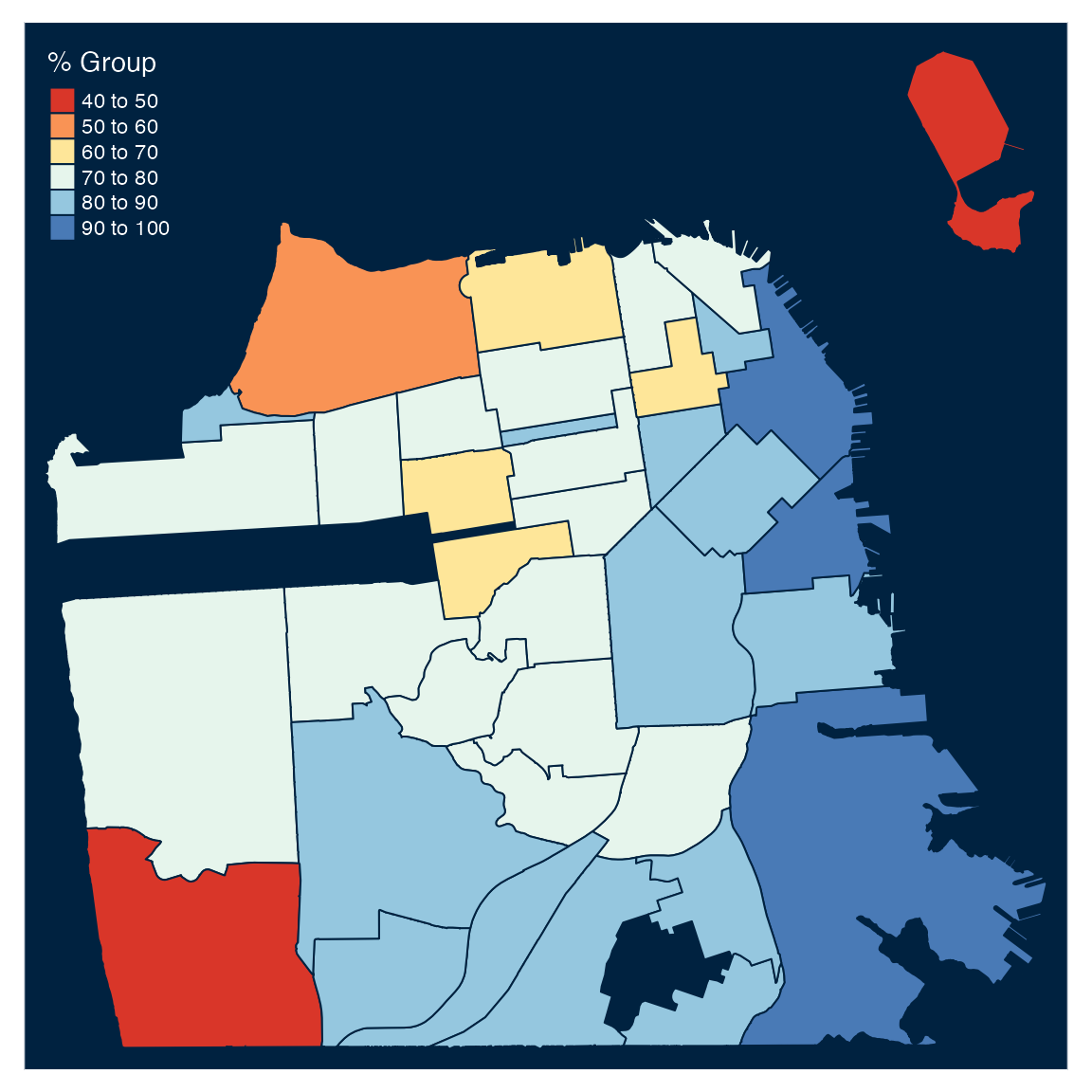
Last but not least, let’s add title and labels for the geographic locations with the tm_text and tm_layout functions:
tm_shape(df) +
tm_polygons("perc_complated",
title = "% Group",
palette = "RdYlBu") +
tm_style("cobalt") +
tm_text("id", size = 0.5) +
tm_layout(
legend.position=c("right", "bottom"),
legend.outside = FALSE,
legend.width = 1,
legend.title.size = 1.2,
legend.text.size = 1,
# legend.outside.size = 0.9,
title= paste("COVID-19 Vaccines Given",
"to San Franciscans by Geography",
sep = " "),
title.position = c("left", "top") ,
inner.margins = c(0.01, .01, .12, .25)) 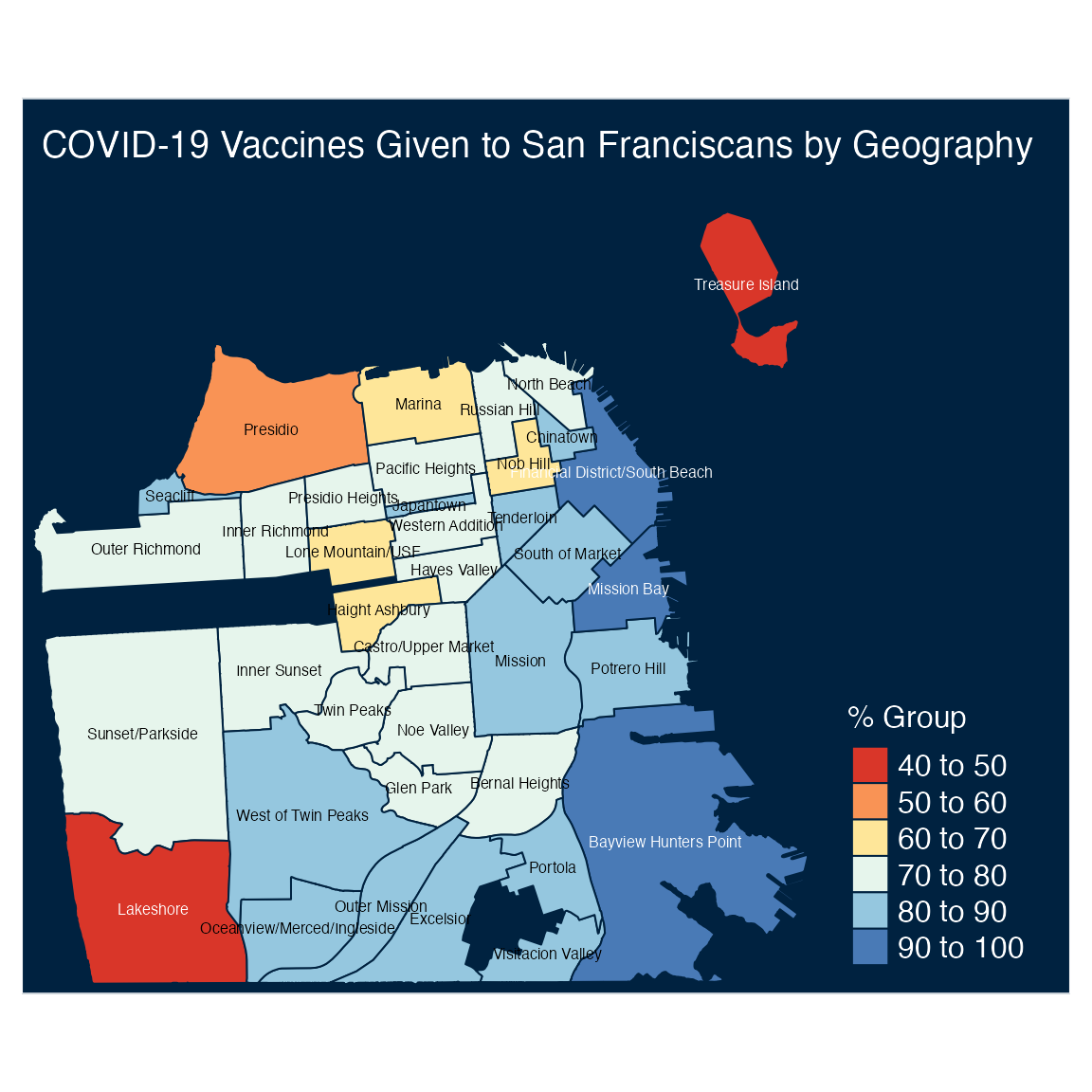
Plotting cases with base plot
The sf package provides a plot method for sf objects (see ?sf:::plot.sf for more information). Similarly to the previews examples above, we will replot the confirmed cases by ZIP code with the plot function:
zip <- covid19sf_geo %>%
dplyr::filter(area_type == "ZCTA") %>%
dplyr::select(count, geometry) %>%
plot(main = "Covid19 Cases by ZIP Code")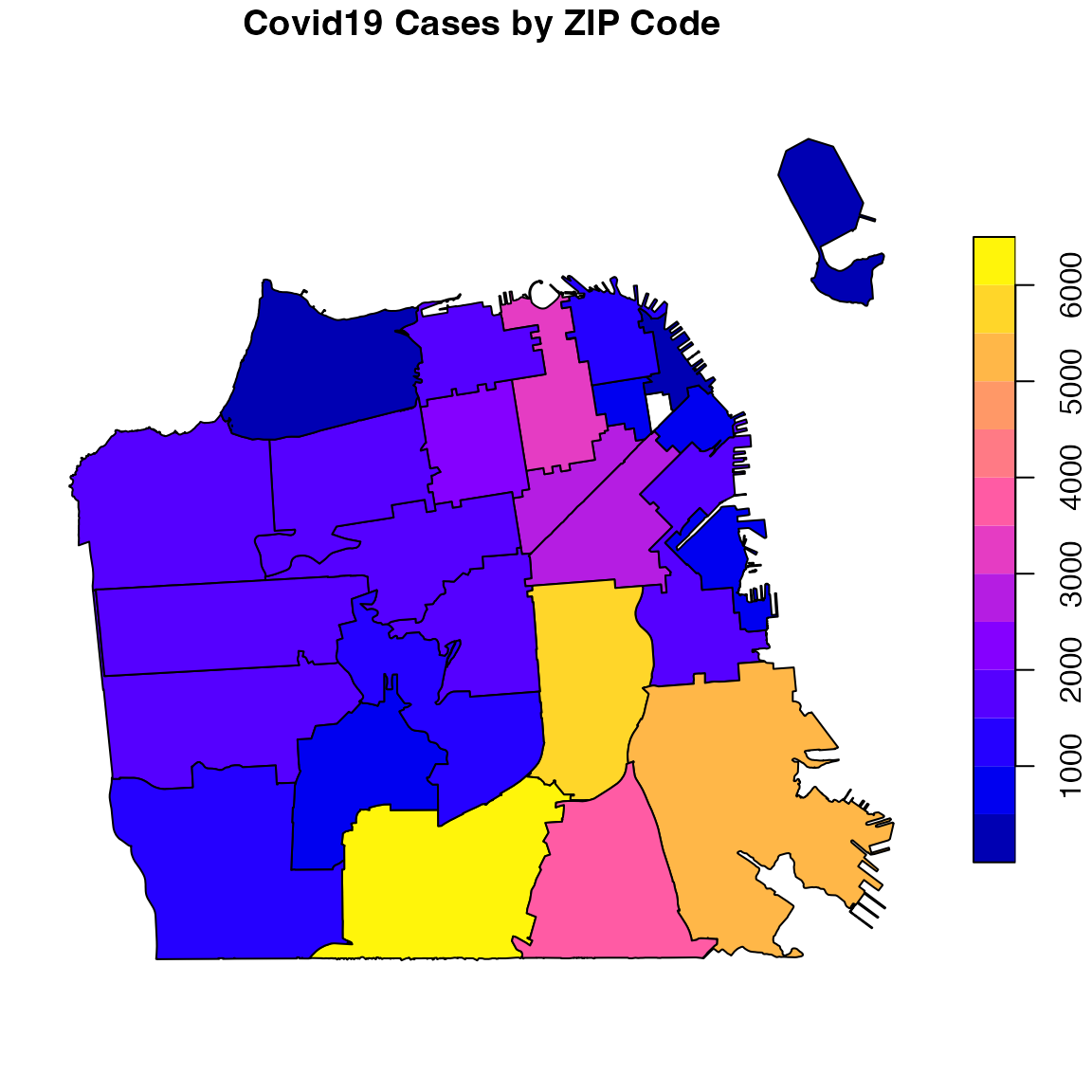
You can define the color palette with the pal argument and set the level of breaks of the color scale by setting the breaks argument to quantile and the number of breaks with the nbreaks argument (which should be aligned with the number of colors on the color palette):
library(RColorBrewer)
pal <- brewer.pal(9, "OrRd")
covid19sf_geo %>%
filter(area_type == "ZCTA") %>%
select(count, geometry) %>%
plot(main = "Covid19 Cases by ZIP Code",
breaks = "quantile", nbreaks = 9,
pal = pal)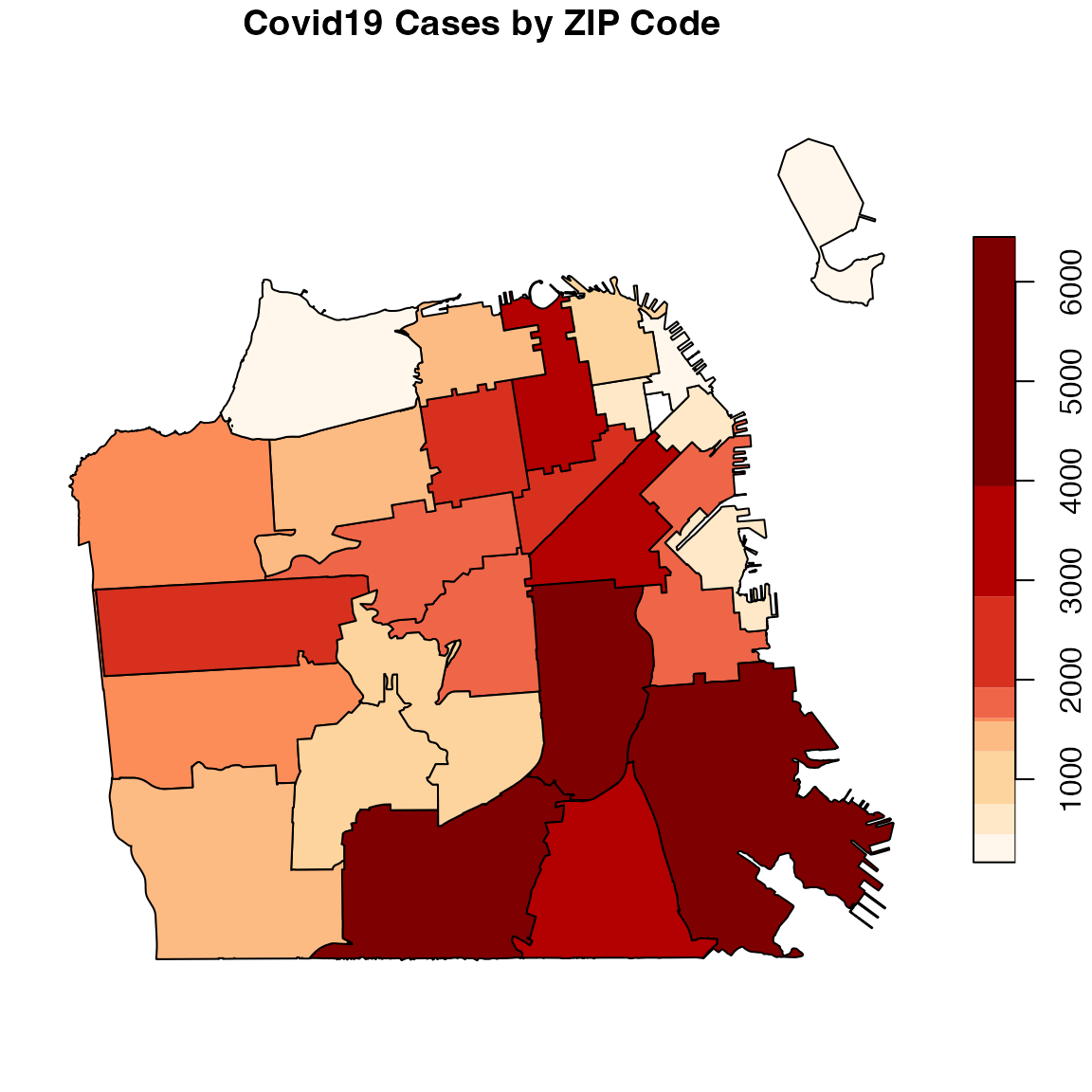
Plotting cases with ggplot2
Plotting sf object can be done with the ggplot2 package natively by using the geom_sf function for plotting sf objects:
library(ggplot2)
covid19sf_geo %>%
filter(area_type == "ZCTA") %>%
ggplot() +
geom_sf(aes(fill=count)) +
ggtitle("Covid19 Cases by ZIP Code")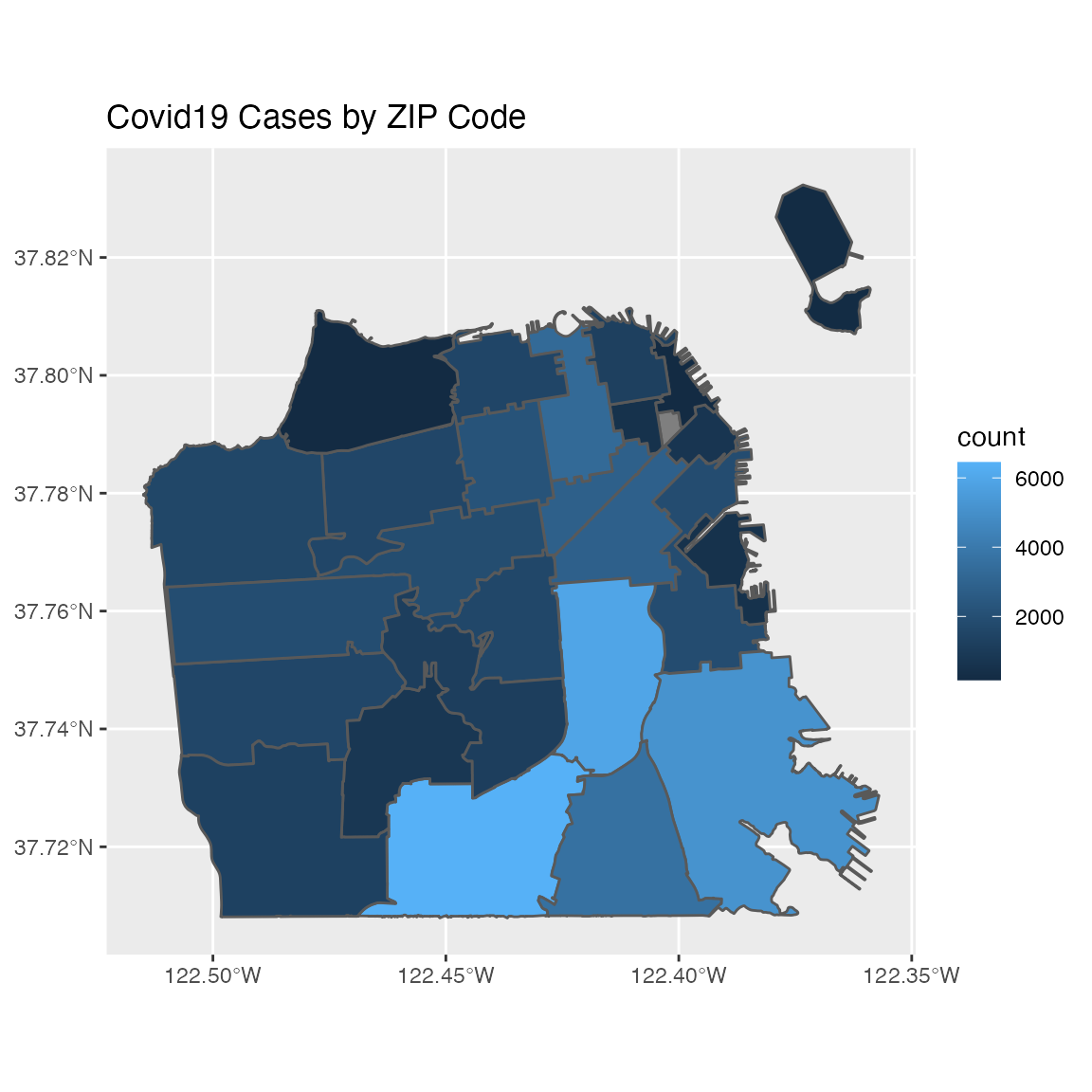
You can customize the polygon color scale by using the scale_viridis function that enables you to select different viridis color palettes. In addition, the geom_sf_label enables you to add labels for each polygon. In the next example, we will replot the count of cases by ZIP code, this time using scale_fill_viridis_b color palette and setting the id variable as the polygon title using the geom_sf_label:
covid19sf_geo %>%
filter(area_type == "ZCTA") %>%
ggplot() +
geom_sf(aes(fill=count)) +
scale_fill_viridis_b() +
geom_sf_label(aes(label = id)) +
ggtitle("Covid19 Cases by ZIP Code")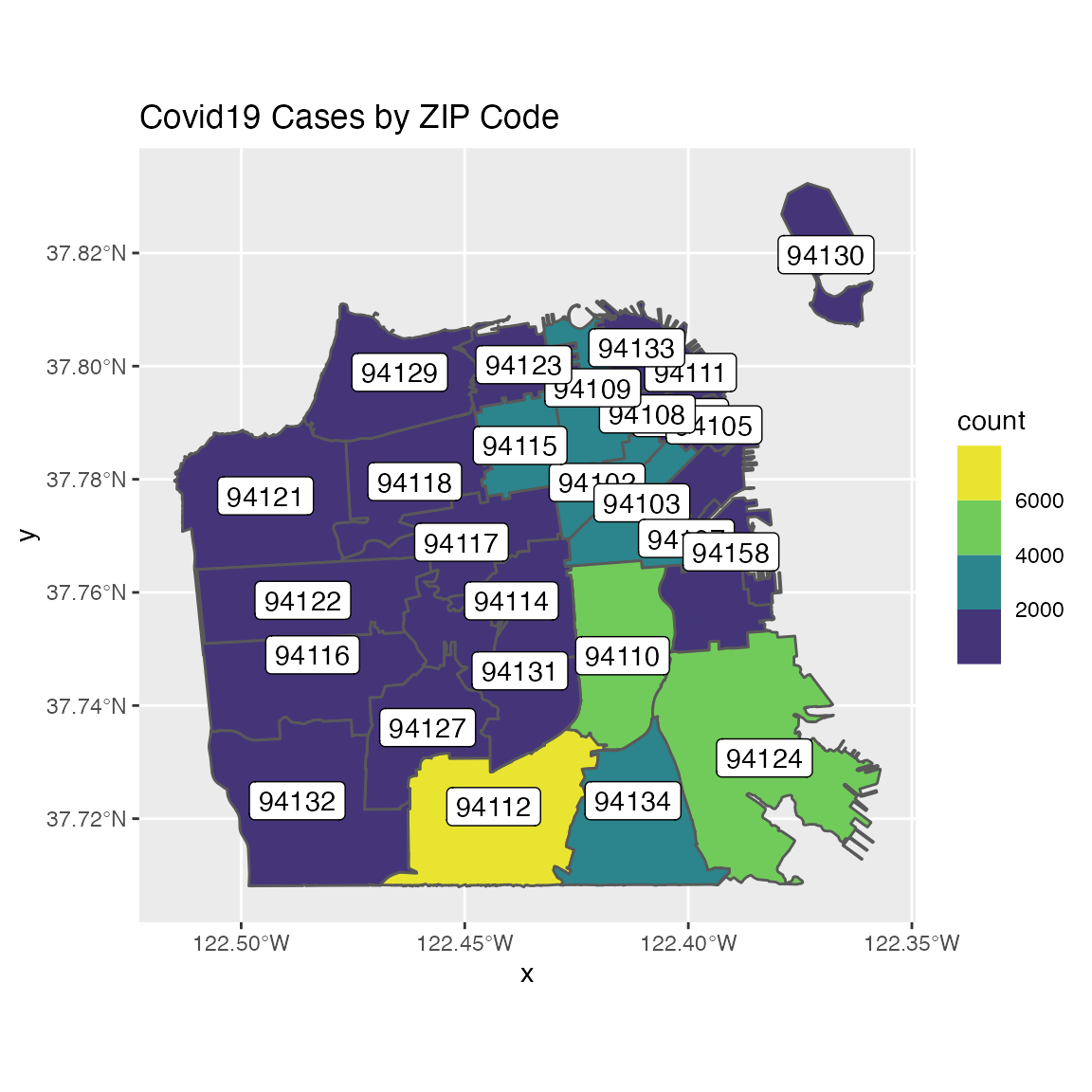
Additional customization of the viridis color palettes can be done by the option argument, where the begin, and end arguments control the color hue:
covid19sf_geo %>%
filter(area_type == "ZCTA") %>%
ggplot() +
geom_sf(aes(fill=count)) +
scale_fill_viridis_b(option = "A",
begin = 0.2,
end = 0.7) +
theme_void() +
ggtitle("Covid19 Cases by ZIP Code")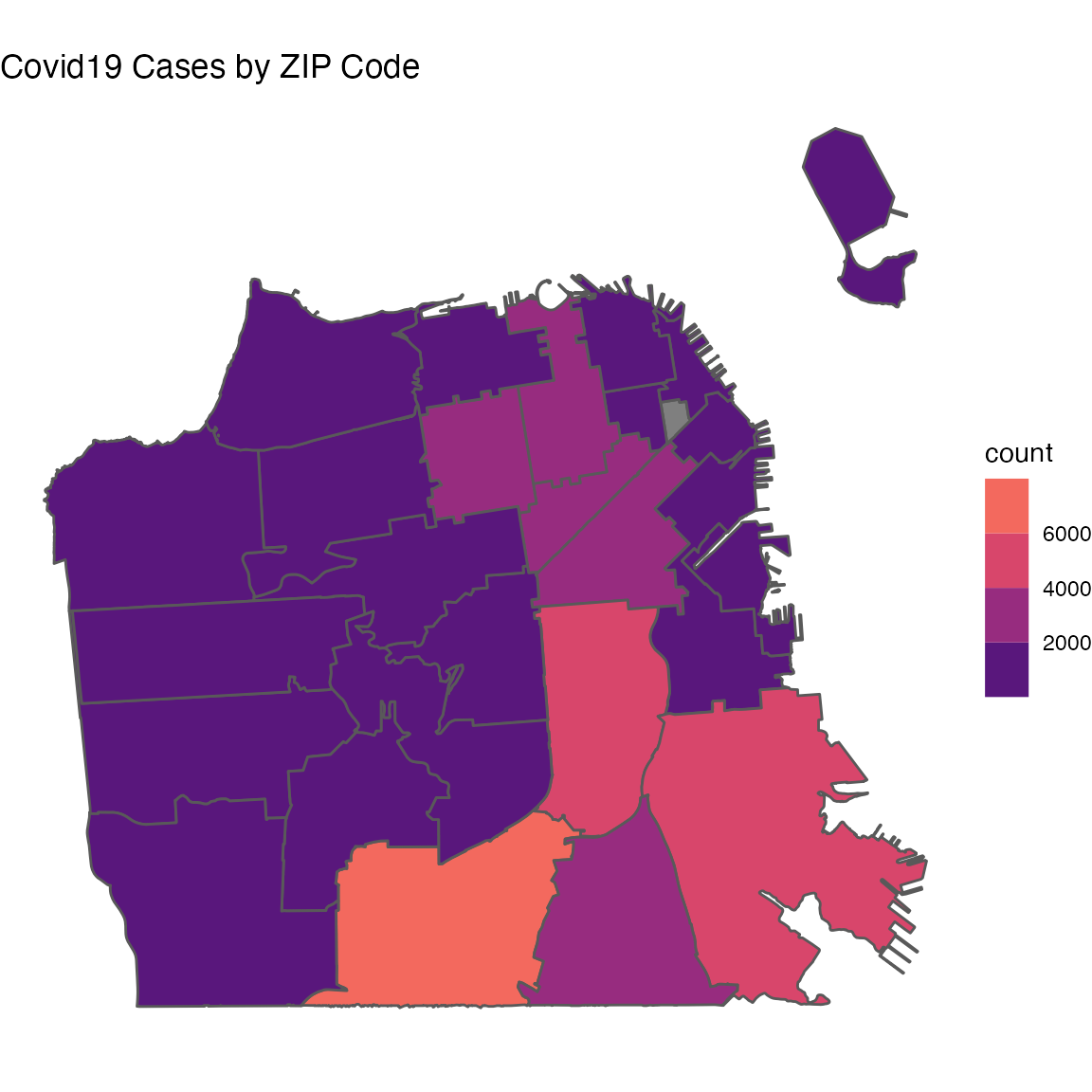
The covid19sf_test_loc dataset
The covid19sf_test_loc datasets provides general metadata about the Covid19 testing locations in San Francisco:
data(covid19sf_test_loc)
head(covid19sf_test_loc)
#> Simple feature collection with 6 features and 16 fields
#> Geometry type: POINT
#> Dimension: XY
#> Bounding box: xmin: -122.4486 ymin: 37.7187 xmax: -122.3849 ymax: 37.78699
#> Geodetic CRS: WGS 84
#> id medical_home
#> 1 6 UCSF
#> 2 1 Carbon Health
#> 3 11 Northeast Medical Services
#> 4 43 SF Department of Public Health in partnership with Color
#> 5 25 SF Department of Public Health in partnership with Color
#> 6 38 <NA>
#> name address phone_number
#> 1 Laurel Heights Campus 3333 California St 4158857478
#> 2 Carbon Health - Castro Clinic 1998 Market St 4157926040
#> 3 San Bruno Ave Clinic 2574 San Bruno Ave 4153919686
#> 4 Alice Griffith 2600 Arelious Walker Dr 311
#> 5 GLIDE Memorial Church 330 Ellis Street 311
#> 6 Test the People by renegade.bio 2730 21st St 5103273540
#> phone_number_formatted testing_hours
#> 1 (415) 885-7478 9am-5pm M-F
#> 2 (415) 792-6040 9am-7pm
#> 3 (415) 391-9686 8:30am-5pm M-F
#> 4 Dial 3-1-1 Saturday, 11/20 only from 10am - 3pm
#> 5 Dial 3-1-1 Tuesdays & Wednesdays, 10am-4pm
#> 6 (510) 327-3540 M, W, and F from 10 AM to 3PM, except holidays
#> popup_or_permanent location_type
#> 1 Pop-Up Private
#> 2 Permanent Private
#> 3 Permanent Private
#> 4 Pop-Up Public
#> 5 Pop-Up Public
#> 6 Pop-Up Private
#> eligibility
#> 1 Must be UCSF patient
#> 2 Must be Carbon Health patient
#> 3 Must be NEMS patient. May register as a new NEMS patient for free
#> 4 Everyone. No insurance required.
#> 5 Everyone. No insurance required.
#> 6 Everyone. No insurance required.
#> cta_text
#> 1 Physician referral required
#> 2 Sign up with provider
#> 3 Sign up with provider
#> 4 Drop in
#> 5 Drop in.
#> 6 Drop in or call ahead for appointments
#> cta_link
#> 1 tel://4158857478
#> 2 https://patient.carbonhealth.com/#/schedule?practiceId=5bdaef44-8ff0-439f-99d7-3285afcc6911&vstId=0f02af4e-1d1c-4a4b-a0f2-e67fca98dd77&vstrId=f743d1d1-d2a1-4fce-a5f9-2e29339b2406&specialtyIds=652c64cc-9fcb-442f-b71e-266390ef2f63
#> 3 https://www.nems.org/covid19/
#> 4 https://sf.gov/GetTestedSF
#> 5 https://sf.gov/GetTestedSF
#> 6 https://www.testthepeople.org/
#> sample_collection_method lab latitude longitude
#> 1 <NA> UCSF <NA> <NA>
#> 2 <NA> Rapid test (Abbot ID NOW) <NA> <NA>
#> 3 <NA> <NA> <NA> <NA>
#> 4 <NA> <NA> 37.7187018 -122.3849448
#> 5 <NA> <NA> 37.7852341 -122.413658
#> 6 <NA> <NA> 37.757629 -122.4094428
#> geometry
#> 1 POINT (-122.4486 37.78699)
#> 2 POINT (-122.4259 37.77001)
#> 3 POINT (-122.4042 37.7291)
#> 4 POINT (-122.3849 37.7187)
#> 5 POINT (-122.4137 37.78523)
#> 6 POINT (-122.4094 37.75763)Plotting the testing locations on map is fairly similar to one of the covid19sf_geo as both are sf objects. The main distinction between the two, is that the covid19sf_test_loc provides the geometry location (e.g., latitude and longitude) as opposed to a polygon. Let’s plot the locations with the mapview package setting the location color by the type (private or public):
Combine cases dist. and testing points
The sync function from the leafsync package enables to combine multiple maps plots. In the following example, we will put side by side the cases split by ZIP code and the testing point in the city map:
m1 <- covid19sf_geo %>%
filter(area_type == "ZCTA") %>%
mapview(zcol = "count",
at = c(0, 200, 400, 800, 1200, 1600, 2000),
col.regions = (c('#fef0d9','#b30000')))
m2 <- covid19sf_test_loc %>% mapview(zcol = "location_type")
sync(m1, m2)