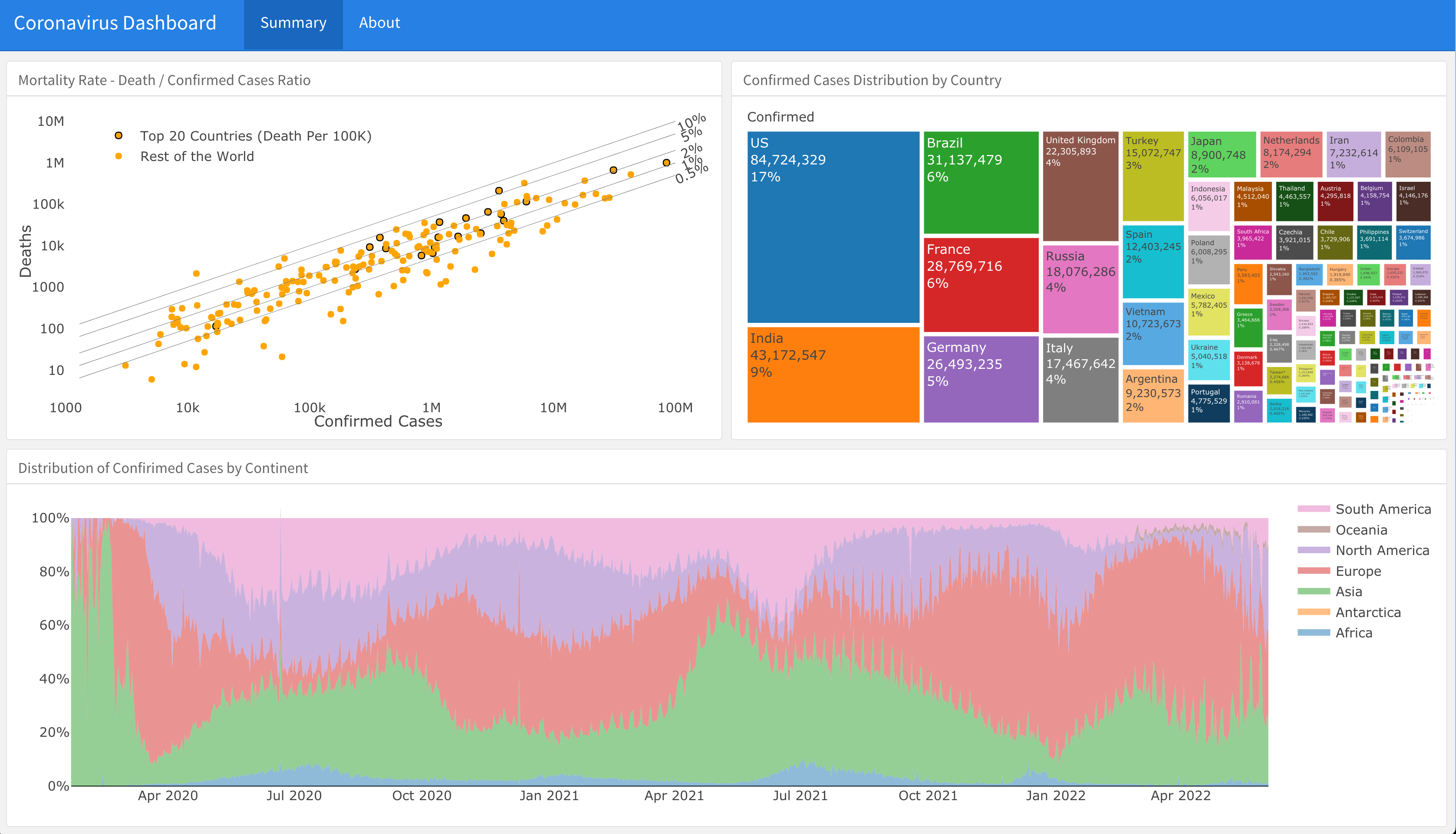
The coronavirus package provides a tidy format for the COVID-19 dataset collected by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University. The dataset includes daily new and death cases between January 2020 and March 2023 and recovery cases until August 2022.
More details available here, and a csv format of the package dataset available here
Data source: https://github.com/CSSEGISandData/COVID-19
Important Notes
- As of March 10th, 2023, JHU CCSE stopped collecting and tracking new cases
- As of August 4th, 2022 JHU CCSE stopped track recovery cases, please see this issue for more details
- Negative values and/or anomalies may occurred in the data for the following reasons:
- The calculation of the daily cases from the raw data which is in cumulative format is done by taking the daily difference. In some cases, some retro updates not tie to the day that they actually occurred such as removing false positive cases
- Anomalies or error in the raw data
- Please see this issue for more details
Installation
Install the CRAN version:
install.packages("coronavirus")Install the Github version (refreshed on a daily bases):
# install.packages("devtools")
devtools::install_github("RamiKrispin/coronavirus")Datasets
The package provides the following two datasets:
-
coronavirus - tidy (long) format of the JHU CCSE datasets. That includes the following columns:
-
date- The date of the observation, usingDateclass -
province- Name of province/state, for countries where data is provided split across multiple provinces/states -
country- Name of country/region -
lat- The latitude code -
long- The longitude code -
type- An indicator for the type of cases (confirmed, death, recovered) -
cases- Number of cases on given date -
uid- Country code -
province_state- Province or state if applicable -
iso2- Officially assigned country code identifiers with two-letter -
iso3- Officially assigned country code identifiers with three-letter -
code3- UN country code -
fips- Federal Information Processing Standards code that uniquely identifies counties within the USA -
combined_key- Country and province (if applicable) -
population- Country or province population -
continent_name- Continent name -
continent_code- Continent code
-
-
covid19_vaccine - a tidy (long) format of the the Johns Hopkins Centers for Civic Impact global vaccination dataset by country. This dataset includes the following columns:
-
country_region- Country or region name -
date- Data collection date in YYYY-MM-DD format -
doses_admin- Cumulative number of doses administered. When a vaccine requires multiple doses, each one is counted independently -
people_partially_vaccinated- Cumulative number of people who received at least one vaccine dose. When the person receives a prescribed second dose, it is not counted twice -
people_fully_vaccinated- Cumulative number of people who received all prescribed doses necessary to be considered fully vaccinated -
report_date_string- Data report date in YYYY-MM-DD format -
uid- Country code -
province_state- Province or state if applicable -
iso2- Officially assigned country code identifiers with two-letter -
iso3- Officially assigned country code identifiers with three-letter -
code3- UN country code -
fips- Federal Information Processing Standards code that uniquely identifies counties within the USA -
lat- Latitude -
long- Longitude -
combined_key- Country and province (if applicable) -
population- Country or province population -
continent_name- Continent name -
continent_code- Continent code
-
The refresh_coronavirus_jhu function enables to load of the data directly from the package repository using the Covid19R project data standard format:
covid19_df <- refresh_coronavirus_jhu()
#> [4;32mLoading 2020 data[0m
#> [4;32mLoading 2021 data[0m
#> [4;32mLoading 2022 data[0m
#> [4;32mLoading 2023 data[0m
head(covid19_df)
#> date location location_type location_code location_code_type
#> 1 2021-12-31 Afghanistan country AF iso_3166_2
#> 2 2020-03-24 Afghanistan country AF iso_3166_2
#> 3 2022-11-02 Afghanistan country AF iso_3166_2
#> 4 2020-03-23 Afghanistan country AF iso_3166_2
#> 5 2021-08-09 Afghanistan country AF iso_3166_2
#> 6 2023-03-02 Afghanistan country AF iso_3166_2
#> data_type value lat long
#> 1 cases_new 28 33.93911 67.70995
#> 2 recovered_new 0 33.93911 67.70995
#> 3 cases_new 98 33.93911 67.70995
#> 4 recovered_new 0 33.93911 67.70995
#> 5 deaths_new 28 33.93911 67.70995
#> 6 cases_new 18 33.93911 67.70995Usage
data("coronavirus")
head(coronavirus)
#> date province country lat long type cases uid iso2 iso3
#> 1 2020-01-22 Alberta Canada 53.9333 -116.5765 confirmed 0 12401 CA CAN
#> 2 2020-01-23 Alberta Canada 53.9333 -116.5765 confirmed 0 12401 CA CAN
#> 3 2020-01-24 Alberta Canada 53.9333 -116.5765 confirmed 0 12401 CA CAN
#> 4 2020-01-25 Alberta Canada 53.9333 -116.5765 confirmed 0 12401 CA CAN
#> 5 2020-01-26 Alberta Canada 53.9333 -116.5765 confirmed 0 12401 CA CAN
#> 6 2020-01-27 Alberta Canada 53.9333 -116.5765 confirmed 0 12401 CA CAN
#> code3 combined_key population continent_name continent_code
#> 1 124 Alberta, Canada 4413146 North America NA
#> 2 124 Alberta, Canada 4413146 North America NA
#> 3 124 Alberta, Canada 4413146 North America NA
#> 4 124 Alberta, Canada 4413146 North America NA
#> 5 124 Alberta, Canada 4413146 North America NA
#> 6 124 Alberta, Canada 4413146 North America NASummary of the total confrimed cases by country (top 20):
library(dplyr)
summary_df <- coronavirus %>%
filter(type == "confirmed") %>%
group_by(country) %>%
summarise(total_cases = sum(cases)) %>%
arrange(-total_cases)
summary_df %>% head(20)
#> # A tibble: 20 × 2
#> country total_cases
#> <chr> <dbl>
#> 1 US 103802702
#> 2 India 44690738
#> 3 France 39866718
#> 4 Germany 38249060
#> 5 Brazil 37076053
#> 6 Japan 33320438
#> 7 Korea, South 30615522
#> 8 Italy 25603510
#> 9 United Kingdom 24658705
#> 10 Russia 22075858
#> 11 Turkey 17042722
#> 12 Spain 13770429
#> 13 Vietnam 11526994
#> 14 Australia 11399460
#> 15 Argentina 10044957
#> 16 Taiwan* 9970937
#> 17 Netherlands 8712835
#> 18 Iran 7572311
#> 19 Mexico 7483444
#> 20 Indonesia 6738225Summary of new cases during the past 24 hours by country and type (as of 2023-03-09):
library(tidyr)
coronavirus %>%
filter(date == max(date)) %>%
select(country, type, cases) %>%
group_by(country, type) %>%
summarise(total_cases = sum(cases)) %>%
pivot_wider(names_from = type,
values_from = total_cases) %>%
arrange(-confirmed)
#> # A tibble: 201 × 4
#> # Groups: country [201]
#> country confirmed death recovery
#> <chr> <dbl> <dbl> <dbl>
#> 1 US 46931 590 0
#> 2 United Kingdom 28783 0 0
#> 3 Australia 13926 115 0
#> 4 Russia 12385 38 0
#> 5 Belgium 11570 39 0
#> 6 Korea, South 10335 12 0
#> 7 Japan 9834 80 0
#> 8 Germany 7829 127 0
#> 9 France 6308 11 0
#> 10 Austria 5283 21 0
#> # … with 191 more rowsPlotting daily confirmed and death cases in Brazil:
library(plotly)
coronavirus %>%
group_by(type, date) %>%
summarise(total_cases = sum(cases)) %>%
pivot_wider(names_from = type, values_from = total_cases) %>%
arrange(date) %>%
mutate(active = confirmed - death - recovery) %>%
mutate(active_total = cumsum(active),
recovered_total = cumsum(recovery),
death_total = cumsum(death)) %>%
plot_ly(x = ~ date,
y = ~ active_total,
name = 'Active',
fillcolor = '#1f77b4',
type = 'scatter',
mode = 'none',
stackgroup = 'one') %>%
add_trace(y = ~ death_total,
name = "Death",
fillcolor = '#E41317') %>%
add_trace(y = ~recovered_total,
name = 'Recovered',
fillcolor = 'forestgreen') %>%
layout(title = "Distribution of Covid19 Cases Worldwide",
legend = list(x = 0.1, y = 0.9),
yaxis = list(title = "Number of Cases"),
xaxis = list(title = "Source: Johns Hopkins University Center for Systems Science and Engineering"))Plot the confirmed cases distribution by counrty with treemap plot:
conf_df <- coronavirus %>%
filter(type == "confirmed") %>%
group_by(country) %>%
summarise(total_cases = sum(cases)) %>%
arrange(-total_cases) %>%
mutate(parents = "Confirmed") %>%
ungroup()
plot_ly(data = conf_df,
type= "treemap",
values = ~total_cases,
labels= ~ country,
parents= ~parents,
domain = list(column=0),
name = "Confirmed",
textinfo="label+value+percent parent")
data(covid19_vaccine)
head(covid19_vaccine)
#> date country_region continent_name continent_code combined_key
#> 1 2020-12-29 Austria Europe EU Austria
#> 2 2020-12-29 Bahrain Asia AS Bahrain
#> 3 2020-12-29 Belarus Europe EU Belarus
#> 4 2020-12-29 Belgium Europe EU Belgium
#> 5 2020-12-29 Canada North America NA Canada
#> 6 2020-12-29 Chile South America SA Chile
#> doses_admin people_at_least_one_dose population uid iso2 iso3 code3 fips
#> 1 2123 2123 9006400 40 AT AUT 40 <NA>
#> 2 55014 55014 1701583 48 BH BHR 48 <NA>
#> 3 0 0 9449321 112 BY BLR 112 <NA>
#> 4 340 340 11589616 56 BE BEL 56 <NA>
#> 5 59079 59078 37855702 124 CA CAN 124 <NA>
#> 6 NA NA 19116209 152 CL CHL 152 <NA>
#> lat long
#> 1 47.5162 14.550100
#> 2 26.0275 50.550000
#> 3 53.7098 27.953400
#> 4 50.8333 4.469936
#> 5 60.0000 -95.000000
#> 6 -35.6751 -71.543000Taking a snapshot of the data from the most recent date available and calculate the ratio between total doses admin and the population size:
df_summary <- covid19_vaccine |>
filter(date == max(date)) |>
select(date, country_region, doses_admin, total = people_at_least_one_dose, population, continent_name) |>
mutate(doses_pop_ratio = doses_admin / population,
total_pop_ratio = total / population) |>
filter(country_region != "World",
!is.na(population),
!is.na(total)) |>
arrange(- total)
head(df_summary, 10)
#> date country_region doses_admin total population continent_name
#> 1 2023-03-09 China NA 1310292000 1404676330 Asia
#> 2 2023-03-09 India NA 1027379945 1380004385 Asia
#> 3 2023-03-09 US 672076105 269554116 329466283 North America
#> 4 2023-03-09 Indonesia 444303130 203657535 273523621 Asia
#> 5 2023-03-09 Brazil 502262440 189395212 212559409 South America
#> 6 2023-03-09 Pakistan 333759565 162219717 220892331 Asia
#> 7 2023-03-09 Bangladesh 355143411 151190373 164689383 Asia
#> 8 2023-03-09 Japan 382415648 104675948 126476458 Asia
#> 9 2023-03-09 Mexico 225063079 99071001 127792286 North America
#> 10 2023-03-09 Vietnam 266252632 90466947 97338583 Asia
#> doses_pop_ratio total_pop_ratio
#> 1 NA 0.9328071
#> 2 NA 0.7444759
#> 3 2.039893 0.8181539
#> 4 1.624368 0.7445702
#> 5 2.362927 0.8910225
#> 6 1.510960 0.7343837
#> 7 2.156444 0.9180335
#> 8 3.023611 0.8276319
#> 9 1.761163 0.7752502
#> 10 2.735325 0.9294048Plot of the total doses and population ratio by country:
# Setting the diagonal lines range
line_start <- 10000
line_end <- 1500 * 10 ^ 6
# Filter the data
d <- df_summary |>
filter(country_region != "World",
!is.na(population),
!is.na(total))
# Replot it
p3 <- plot_ly() |>
add_markers(x = d$population,
y = d$total,
text = ~ paste("Country: ", d$country_region, "<br>",
"Population: ", d$population, "<br>",
"Total Doses: ", d$total, "<br>",
"Ratio: ", round(d$total_pop_ratio, 2),
sep = ""),
color = d$continent_name,
type = "scatter",
mode = "markers") |>
add_lines(x = c(line_start, line_end),
y = c(line_start, line_end),
showlegend = FALSE,
line = list(color = "gray", width = 0.5)) |>
add_lines(x = c(line_start, line_end),
y = c(0.5 * line_start, 0.5 * line_end),
showlegend = FALSE,
line = list(color = "gray", width = 0.5)) |>
add_lines(x = c(line_start, line_end),
y = c(0.25 * line_start, 0.25 * line_end),
showlegend = FALSE,
line = list(color = "gray", width = 0.5)) |>
add_annotations(text = "1:1",
x = log10(line_end * 1.25),
y = log10(line_end * 1.25),
showarrow = FALSE,
textangle = -25,
font = list(size = 8),
xref = "x",
yref = "y") |>
add_annotations(text = "1:2",
x = log10(line_end * 1.25),
y = log10(0.5 * line_end * 1.25),
showarrow = FALSE,
textangle = -25,
font = list(size = 8),
xref = "x",
yref = "y") |>
add_annotations(text = "1:4",
x = log10(line_end * 1.25),
y = log10(0.25 * line_end * 1.25),
showarrow = FALSE,
textangle = -25,
font = list(size = 8),
xref = "x",
yref = "y") |>
add_annotations(text = "Source: Johns Hopkins University - Centers for Civic Impact",
showarrow = FALSE,
xref = "paper",
yref = "paper",
x = -0.05, y = - 0.33) |>
layout(title = "Covid19 Vaccine - Total Doses vs. Population Ratio (Log Scale)",
margin = list(l = 50, r = 50, b = 90, t = 70),
yaxis = list(title = "Number of Doses",
type = "log"),
xaxis = list(title = "Population Size",
type = "log"),
legend = list(x = 0.75, y = 0.05))Dashboard
Note: Currently, the dashboard is under maintenance due to recent changes in the data structure. Please see this issue
A supporting dashboard is available here
Data Sources
The raw data pulled and arranged by the Johns Hopkins University Center for Systems Science and Engineering (JHU CCSE) from the following resources:
- World Health Organization (WHO): https://www.who.int/
- DXY.cn. Pneumonia. 2020. https://ncov.dxy.cn/ncovh5/view/pneumonia.
- BNO News: https://bnonews.com/index.php/2020/04/the-latest-coronavirus-cases/
- National Health Commission of the People’s Republic of China (NHC):
http:://www.nhc.gov.cn/xcs/yqtb/list_gzbd.shtml
- China CDC (CCDC): http:://weekly.chinacdc.cn/news/TrackingtheEpidemic.htm
- Hong Kong Department of Health: https://www.chp.gov.hk/en/features/102465.html
- Macau Government: https://www.ssm.gov.mo/portal/
- Taiwan CDC: https://sites.google.com/cdc.gov.tw/2019ncov/taiwan?authuser=0
- US CDC: https://www.cdc.gov/coronavirus/2019-ncov/index.html
- Government of Canada: https://www.canada.ca/en/public-health/services/diseases/2019-novel-coronavirus-infection/symptoms.html
- Australia Government Department of Health:https://www.health.gov.au/health-alerts/covid-19
- European Centre for Disease Prevention and Control (ECDC): https://www.ecdc.europa.eu/en/covid-19/country-overviews
- Ministry of Health Singapore (MOH): https://www.moh.gov.sg/covid-19
- Italy Ministry of Health: https://www.salute.gov.it/nuovocoronavirus
- 1Point3Arces: https://coronavirus.1point3acres.com/en
- WorldoMeters: https://www.worldometers.info/coronavirus/
- COVID Tracking Project: https://covidtracking.com/data/. (US Testing and Hospitalization Data. We use the maximum reported value from “Currently” and “Cumulative” Hospitalized for our hospitalization number reported for each state.)
- French Government: https://dashboard.covid19.data.gouv.fr/
- COVID Live (Australia): https://covidlive.com.au/
- Washington State Department of Health:https://doh.wa.gov/emergencies/covid-19
- Maryland Department of Health: https://coronavirus.maryland.gov/
- New York State Department of Health: https://health.data.ny.gov/Health/New-York-State-Statewide-COVID-19-Testing/xdss-u53e/data
- NYC Department of Health and Mental Hygiene: https://www.nyc.gov/site/doh/covid/covid-19-data.page and https://github.com/nychealth/coronavirus-data
- Florida Department of Health Dashboard: https://services1.arcgis.com/CY1LXxl9zlJeBuRZ/arcgis/rest/services/Florida_COVID19_Cases/FeatureServer/0 and https://fdoh.maps.arcgis.com/apps/dashboards/index.html#/8d0de33f260d444c852a615dc7837c86
- Palestine (West Bank and Gaza): https://corona.ps/details
- Israel: https://govextra.gov.il/ministry-of-health/corona/corona-virus/
- Colorado: https://covid19.colorado.gov/